4.10 LAUNCH MENU
- 4.10.1 about the launch menu
- 4.10.2 populating the launch menu
- 4.10.3 supported external programs
- 4.10.4 auto-installed external programs (Clustal, Chromas, t-coffee)
 4.10.1 about the launch menu
4.10.1 about the launch menu
SEQtools depends on a number of external programs. The important resources provided by NCBI for example
are all included in the SEQtools setup file and are available in the auxuliary8.exe update file. Other programs come
with their own setup file and must be installed as individual programs for SEQtools to use them (GeneDoc, TreeView).
It is therefore necesseay to inform SEQtools where these programs are located. This is done as described in
the popularing the launch menu below. It is possible to access the programs directly from
the Launch menu.
Other external programs (Chromas, T-Coffee) have no specific SEQtools interface but are used - and installed - by the
setup file.
Finally you may wish to have easy access to certain other programs. This can be achieved by including the path to such
programs on the Launch menu.
 4.10.2 populating the launch menu
4.10.2 populating the launch menu
Adding new items to the Launch menu is described in detail on
the preferences menu.
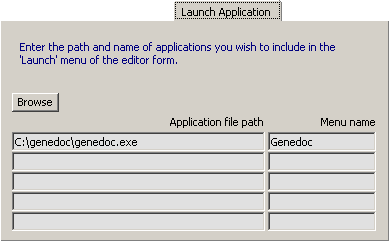
Briefly, select the line where you want the program to appear, browse to the location of the exe file of the program
and press <Enter> when you have have found it. The application title is automatically inserted in the editable right
text field. Remember this is only possible after the application is properly installed on your pc. A maximum
of five external applications can be included.
 4.10.3 supported external programs
4.10.3 supported external programs
Two separately installed external programs (GeneDoc, TreeView) are interfaced to SEQtools and are accessed from the Clustal results form. A third external program,T-Coffee, is a dos program, installed by the SEQtools setup file and runs in the background without a user interface.
All three programs are used to postprocess sequence aligments created by ClustalW. To use these programs, first perform a
sequence alignment with ClustalW. When the output format is set to GCG - and only then - the three programs can be
accessed from the main menu of the Clustal results form.

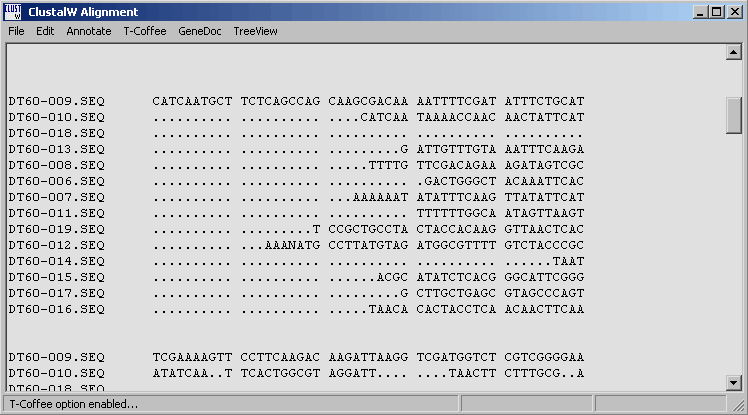
The ClustalW results form with the three external programs on the menu line. Note that the results
menu will only display these menu items if the ClustalW output is set to GCG before the alignment process
is launched from the alignment form above.
 4.10.3.1 GeneDoc - alignment editor
4.10.3.1 GeneDoc - alignment editor
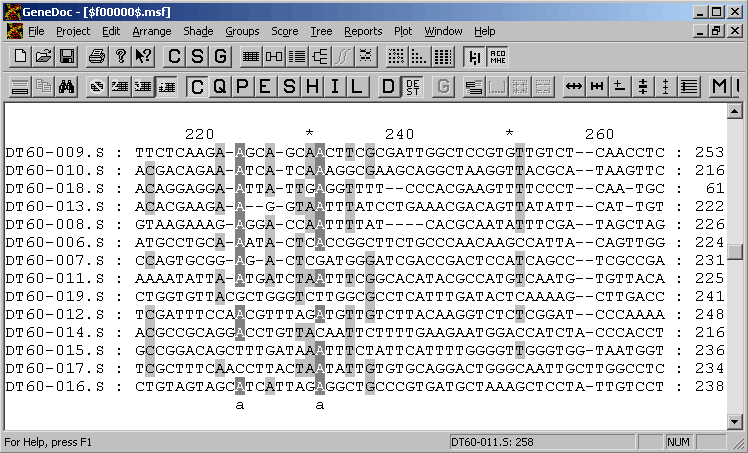
GeneDoc is a very powerful editor with numerous options for editing and annotating sequence alignments. It will take some experimentation to get familiar with this exellent program. The program is written byKarl Nicholasand is freeware. Download the program from Karl Nicholas' homepage.
The GeneDoc editor in action:
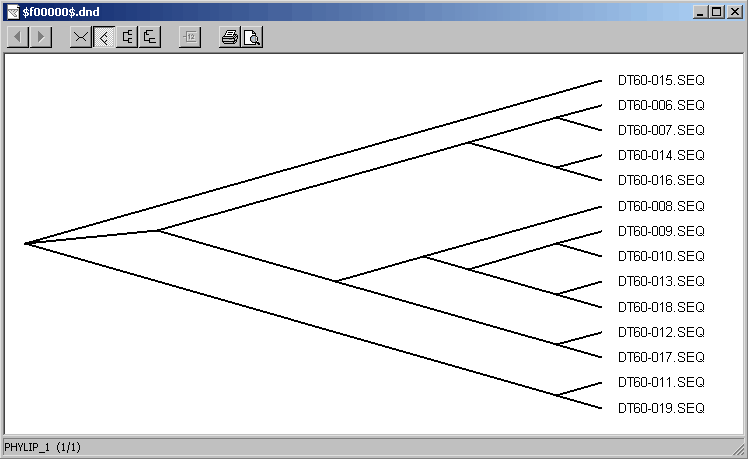
 4.10.3.2 TreeView - draws phylogenetic trees
4.10.3.2 TreeView - draws phylogenetic trees
TreeView is written by Roderic D. M. Page and is freeware. You will find amanualto TreeView at the Glascow University homepage. The program can bedownloadedat the same place.
Phylogenetic tree creayed by TreeView for the set of sequences shown in the GeneDoc editor above:
 4.10.4 auto-installed external programs
4.10.4 auto-installed external programs
ClustalW,ClustalX,Chromasas well ast-coffeeare all included in the SEQtools setup file and are installed automatically. Read more about these programs on their respective homepages.
© 2002-2010S.W. Rasmussen (revised: )