4.14 PREFERENCES MENU
- 4.14.1 about preferences
- 4.14.2 general settings
- 4.14.2.1 project files
- 4.14.2.2 colors and fonts
- 4.14.2.3 launch application
- 4.14.2.4 launch url
- 4.14.2.5 backup settings
- 4.14.2.6 checksum calculation
- 4.14.2.7 dos directory
- 4.14.2.8 footnotes
- 4.14.3 project settings
- 4.14.3.1 project title
- 4.14.3.2 user data
- 4.14.3.3 sequence format
- 4.14.3.4 color patterns, nucleotide
- 4.14.3.5 color patterns, protein
- 4.14.3.6 color patterns, primer
- 4.14.3.7 header
- 4.14.3.8 trace file folder
- 4.14.3.9 global timeout
- 4.14.3.10 project blast settings
- 4.14.3.11 backup settings
- 4.14.4 form behavior settings
- 4.14.5 description line format settings
- 4.14.6 chromatogram import settings
- 4.14.6.1 fixed schemes
- 4.14.6.2 options for basecalling
- 4.14.6.3 trimming raw sequences
- 4.14.6.4 n-threshold
- 4.14.6.5 gap-quality
- 4.14.7 ncbi settings, firewall
- 4.14.8 internet connection and server settings
- 4.14.9 compose search data file
- 4.14.10 log- and ini-file viewer
- 4.14.11 application color coding
 4.14.1 about preferences
4.14.1 about preferences
SEQtools contains various options for controlling program behavior grouped
under the Preferences menu of the main editor form.
When you exit SEQtools, the settings are saved in an ini-file located in the
application folder as a plain text file. Each instance of SEQtools gets its own ini-file
making it possible to set different preferences for different instances of SEQtools. For
example you can set different sequence color pattern and background color for instances
for handling nucleotide, protein and primer sequences. Read more
aboutcommand line optionsin
section 3.3 Features of the manual.
It is recommended that you go through and set program preferences as you prefer the first
time you launch SEQtools after installing the program. Pay special attention to settings
for internet connection and the NCBI ini-file. The latter setting varies depending on
your operating system. (Windows2000 or Windows XP)
The different groups of preferences are described in some detail below. Quite a number of
user questions relates to problems caused by incorrect preference settings so please read
this section carefully.
 4.14.2 general settings
4.14.2 general settings
The eight tabs of the general settings form are displayed below. In most cases the effect of changing these
settings are self explanatory.
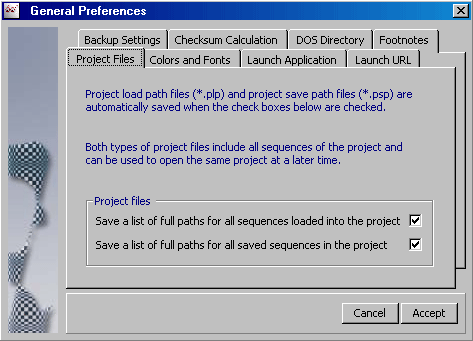
4.14.2.1 Project Files - Enables auto-saving plp and psp files each time a project is saved.
This implies that you can re-open the project either with the sequences originally loaded into the project
(*.plp) or with the files included
after editing and saving the project (*.psp).
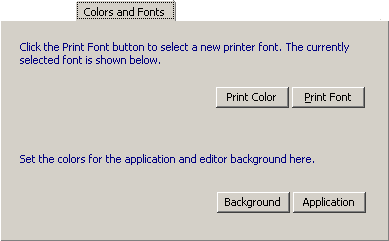
4.14.2.2 Colors and fonts - SEQtools requires that you use fonts with fixed character-spacing such
as Courier New or Monospace (included in the SEQtools setup file). Using fonts with variable character
width will cause the sequence to be distorted when displayed in the editor and in the various sequence lists. In most
cases, however, SEQtools uses hard coded font which cannot be changes by the user.
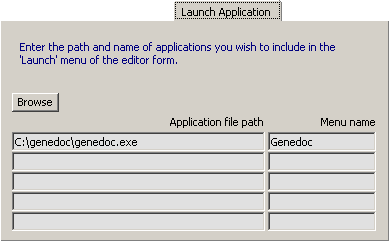
4.14.2.3 Launch Application - This tab allows you to populate the Launch menu of the main editor with up
to five applications which will be launched by clicking the menu item. To add an application to the list place the cursor
on the line where you wish the application to appear. Click Browse and navigate to the exe-file of the application
be to included. Clicking the exe-file places the full path in the left field and the title of the exe-file in the right.
Edit the title of the exe-file if you wish.
4.14.2.4 Launch URL - On this tab you can edit the default list of web resources: Type the url of the website in
the left text field and the title of the site in the right field. Maximum number of urs is 10. URL incuding the word NCBI
will be identified by an NCBI icon in the menu item, other url with a globe.
4.14.2.5 Backup Settings - Setting the interval between timed backups to a positive value specifying the number
of minutes between auto-backups activates the auto-backup function. It is not recommended to use this function when you
are working with large projects (> 300 sequences). SEQtools will notify you if the size of the projects exceeds 300
and offer to turn off auto-backup.
Note that the batch blast function - which may run over several days - has a separate function for timed saving rearch
results to avoid loss of data in case of power failure during the batch job. This tab is also included in
the Project Settings form.
4.14.2.6 Checksum Calculation - This tab controls the checksum function. You may gain some speed improvement
by turning this function off.
4.14.2.7 DOS Directory - By default the special directory for the various external programs used in SEQtools
to perform certain tasks is the one given in the tab below. It is possible - but not recommended - to change this path.
I have not tested the effect of placing the dos folder on a different drive and any experiments is at your
own risk.
If you change the path SEQtools moves the content to the new destination when you click Accept.
4.14.2.8 Footnotes - In some cases it may be convenient to include a footer when you save a sequence file.
 4.14.3 project settings
4.14.3 project settings
The settings included in this form affect various aspects of the current sequence. .
4.14.3.1 Project Titles - This tab is displayed before loading of sequences into the project commences. It furthermore
includes a checkbox activating validation of sequences while loading. This option which is only relevant when you are
loading non-SEQtools formatted sequences slows the loading process somewhat down.
4.14.3.2 User Data - This tab enables you to personalise your project by entering your name and
e-mail address.
4.14.3.3 Project Files - The tab allows you to set the display preferences for sequence formatting, i.e., whether
or not to use upper, lower or both cases; to allow spaces in the displayed sequence and to set line and block length and
line spacing.
4.14.3.4 Color Patterns, nucleotide - You can change the color of the nucleotides in nucleotide sequences.
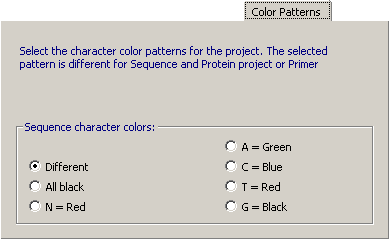
4.14.3.5 Color Patterns, protein - You can change the color of individual residues in protein sequences with
the options included in this tab. Note you can selectively highlight amino acids according to their properties with
the Basic, Hydrophilic, Acidic and Hydrophobic options.

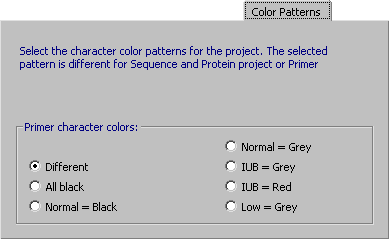
4.14.3.6 Color Patterns, primer - You can change the color of individual residues of primer sequences with the
options included in this tab. Note that you can selectively highlight degenerate positions in the sequence with
the IUB = Red option.
4.14.3.7 Header - Note that thecompositionof the
sequence header is controlled by a separate set of quite extensive options which allows you to precisely select which part
of the total sequence annotation you wich to include in the displayed header.
4.14.3.8 Trace File Folder - This tab allows you to set the path to the folder containing the chromatograms from
which sequences in the project are derived. The path is automatically set to that of imported/basecalled chromatograms
(=trace files) when they are loaded. If, however, you import chromatograms located in different folders it is necessary
to set the path to the folder containing the underlying trace files. Obviously it is preferrable to place all trace
files to be included in a project in the same folder.

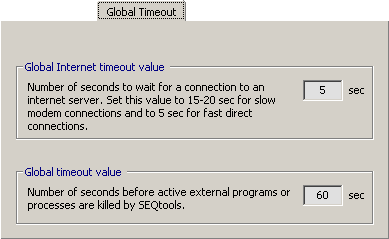
4.14.3.9 Global Timeout - SEQtools uses a number of external programs running under the dos operating system.
To do this SEQtools creates a "memory space" to hold the process and monitors the process to detect when it
ends, i.e., when the task is completed. If something goes wrong and the externat program fails to issue the completed
signal the process will run for ever unless a timeout is set to kill the process if no response is received within a
certain period of time.
If everything works as it should the timeout value has no effect as long as it is sufficiently large to allow external
programs to complete. If, on the other hand, the problem is temporary, a long time out value may allow the halted process
to resume and run to completion. You need to experiment to find the optimal value. Sixty seconds works fine in most
cases.
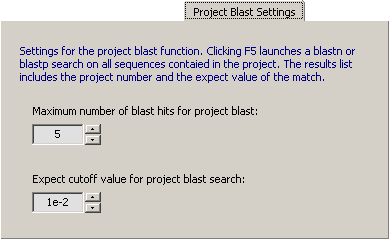
4.14.3.10 Project Blast Settings - Pressing <F5> will create a local database containing all sequences on
the project and perform a blast search on this database, i.e., perform a Project Blast. The result of this project
blast search is displayed in the sequence list in the right hand panel of the main editor (on a light blue background).
The settings in this tab controls the number of matches to display and an expect value cutoff. Setting the maximum number of
matches to zero turns off this function (which there is no reason to do).
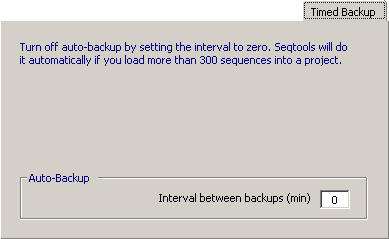
4.14.3.11 Backup Settings - Setting the interval between timed backups to a positive value specifying the number
of minutes between auto-backups activates the auto-backup function. It is not recommended to use this function when you
are working with large projects (> 300 sequences). SEQtools will notify you if the size of the projects exceeds 300
and offer to turn off auto-backup.
Note that the batch blast function - which may run over several days - has a separate function for timed saving search
results to avoid loss of data in case of power failure during the batch job. Note that a Backup Settings tab is also
included in the General Settings form.
 4.14.4 form behavior settings
4.14.4 form behavior settings
SEQtools uses separate forms for most tasks. This often results in the screen being crowded with forms -
the one you which to use in most case being hidden behind other forms. To give you the option to excert some control
of the form behavior the settings in the Form-Stay-On-Top preferences may be useful. By putting a checkmark in
the box for the form the form will remain at the top position, even when it hasn't got the focus.
Setting these preferences requires some experimenting...

 4.14.5 description line format settings
4.14.5 description line format settings
One of the strong features of SEQtools is the functions for customising sequence listing to facilitate
getting an overwies of the sequences contained in the project.
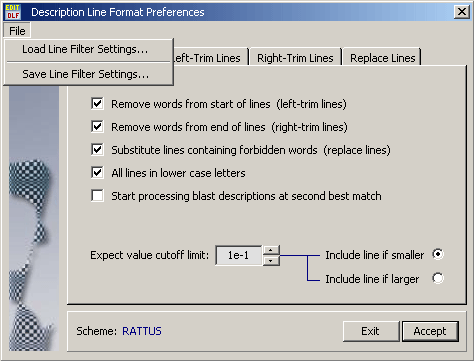
The four tabs included in Description Line Format settings are describes below.
4.14.5.1 Format Settings - The check boxes on this tab enable/disable different formatting options. In addition it is
possible to set a cutoff expect value to exclude blast results with worse than the specified match from the
sequence list.
The File menu includes save and load options for the selected formatting scheeme to allow you to easily
switch between different formatting scheemes. Line formatting scheeme files (*.lfs) are saved in the DataFiles
folder of the main SEQtools application folder. A few *,lfs files are included in the setup file to illustrate how
quite detailed line formatting can be acomplished with the different line formatting options.
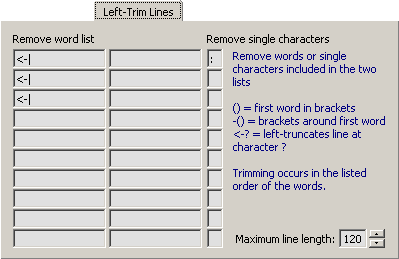
4.14.5.2 Left-Trim Lines - With this option you can enter a list of words to be removed from the start of
the description lines. Removing occurs sequentially from the top of the list.
Assume for example that a large number of the description lines have the word Rat as the first word
hypothetical as the second word. Adding Rat as the first word in the list and hypothetical
as the second will the remove both words from the description lines. Whereas the two words in reverse order will
remove nothing from such lines.
In some cases you may want to remove everything from a specific character. This is possible by adding "<-"
in front of the character from which you want to truncate the line. The"()" code removes the first word in
brackets whereas "-()" removes only the brackets. Single characters can be removed by adding the character
in leftmost column.
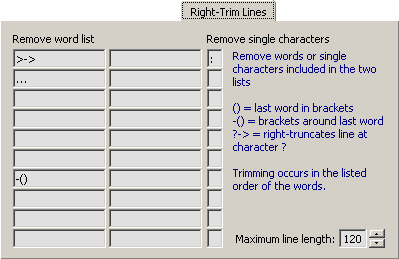
4.14.5.3 Right-Trim Lines - This tab includes the same options as the Left-Trim Lines only the formatting
occurs from the right end of the lines.
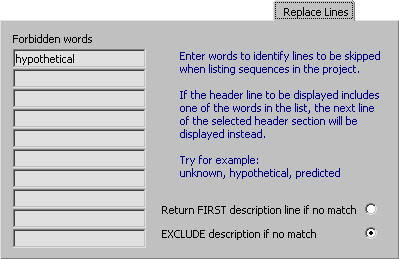
4.14.5.4 Replace Lines - In many cases the best match of a blast search returns a description line indication
that the match is to a hypothetical, putative, similar to, deduced or
a probable protein. This is not very helpful if you are looking for an indication of a possible function of
the protein.
By using the Replace Line filter you kan make SEQtools skip description lines containing the forbidden
words and instead look for alternatives further down the blast results - but still within the expect value cutoff set at the
first tab of this form.
If SEQtools is unable to find a description line without one of the forbidden words and within
the expect value limit the first description line (=best match) is returned.
 4.14.6 chromatogram import settings
4.14.6 chromatogram import settings
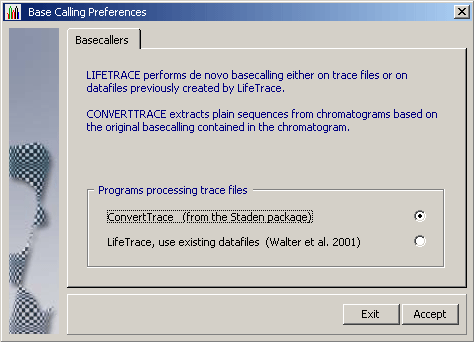
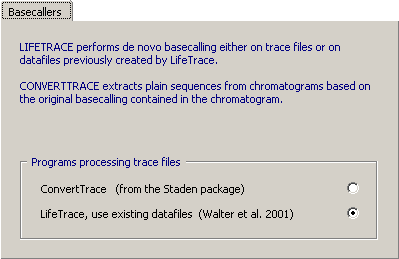
The behavior of ConvertTrace is handled by SEQtools and do not require any user
intervention. The format of trace files to be imported is automatically detected by SEQtools and passed on to
ConvertTrace. ConvertTrace accepts most common trace file formats including ABI, ABD and SCF.
The SEQtools interface to LifeTrace on the other hand gives the user extensive control over the basecalling process
as well as of the removal of low quality regions from raw sequences. Read more about basecalling with LifeTrace under the File menu.
4.14.6.1 Fixed scheemes - The SEQtools user interface to LifeTrace currently includes three
fixed schemes for basecalling and trimming trace files (sloppy, standard and stringent), all based on:
- - a minimum qscore value causing SEQtools to insert a N in the sequence instead of an ambiguous base
- - the size of a sliding window in which none of the bases have qscore values below a given cutoff value
- - the minimum accepted qscore in the sliding window
- - a minimum gap quality score causing SEQtools to print the two bases flanking the gap in lower case characters.
4.14.6.2 Options for Basecalling - Setting optimal parameters for basecalling with LifeTrase is not trivial
and may require some experiments before the best result is achieved. Useful hints are described below.
The currently active parameters for basecalling and trimming are shown in a text box on the Preset
Options tab.
To compensate for differences in signal strength between individual sequence runs, an option is
included to slightly modify the parameters compensating for varying signal strength among different trace files.
With this function, Adaptive N-Threshold, enabled qscores are adjusted in the following way:
QualityScore = QualityScore + ((MeanScore - NeutralMean) / 15).
As can be seen from the equation above, this adjustment has no effect when the mean of all quality scores for the trace
file is equal to the 'neutral mean'. For mean scores greater than the 'neutral mean', quality score cutoffs are increased
while score cutoffs smaller than the 'neutral mean' score are reduced.
This option allows you to introduce a slight adjustment of the set parameters to compensate for small variations in the
overall quality of individual trace files.
This option as well as the quality scores, window sizes and gap quality scores currently selected for the three fixed
settings may be adjusted if suggested by further testing of the function.
4.14.6.3 Trimming Raw Sequences - Based on qscore values and a set sliding window length it is possible
to perform a quite precise trimming of raw sequence files. Activate the adaptive N-threshold option to reduce
the effect of varying peak height in the chromatograms.
4.14.6.4 N-Threshold - Options to control when the basecaller inserts an N when the basecalling is ambiguous.
Activate the adaptive N-threshold option to reduce the effect of varying peak height in the chromatograms
4.14.6.5 Gap-Quality - Finally it is possible to have flanking, dubious bases displayed in lower case when the peak spacing is irregular (gap_qscore). This information is relevant to highlight regions where the number of bases within a short region of the sequence cannot be determined precisely.
 4.14.7 ncbi settings, firewall
4.14.7 ncbi settings, firewall
NCBI Ini-File - Using the various programs provided by NCBI require that a NCBI ini-file is present in the Windows folder.
The ini-file contains the path to the NCBI data folder. Depending on your operation system, the name of the Windows
folder is WINNT (Windows 2000) or Windows (Windows XP). This difference must be incorporated in the NCBI ini-file.
In case you are behind a firewall you may also have to edit this information in the NCBI ini-file.
The NCBI Ini-File editor include relevant information and options for these tasks.
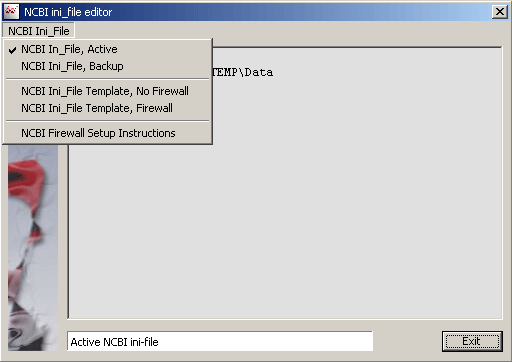
 4.14.8 internet connection and server settings
4.14.8 internet connection and server settings
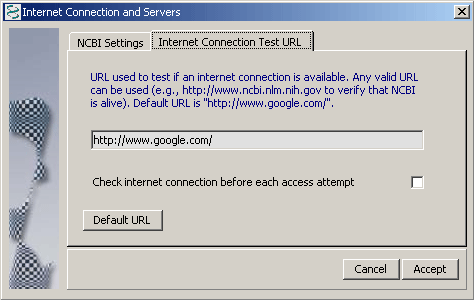
4.14.8.1 Internet Connection - SEQtools verifies the internet connection each time the program is launced. This is done by connection to the Google server at the URL http://www.google.com/. It is possible to change this URL if you wish to test the connection to a different server.
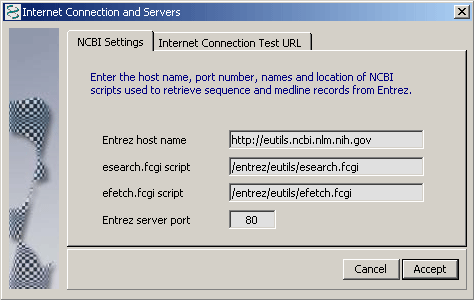
4.14.8.2 NCBI Resources - The NCBI Settings tab includes URLs to various NCBI resourcers. These values should not be changed by the user (unless you known exactly what you are doing).
 4.14.9 compose search data file
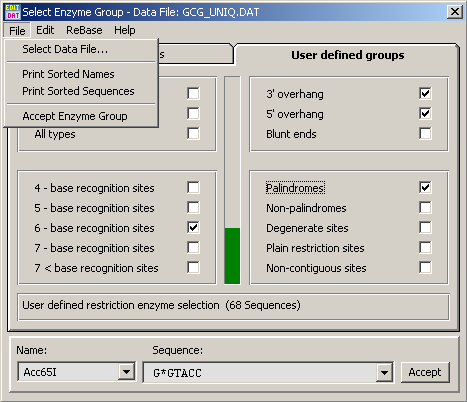
4.14.9 compose search data file
The default project restriction enzyme data file can be composed using the enzyme group editor shown below. The first tab includes a number of fixed patterns while the second tab allows you to create a highly specific enzyme file.
After clicking Accept the selected enzyme file will be the default until it is changed.

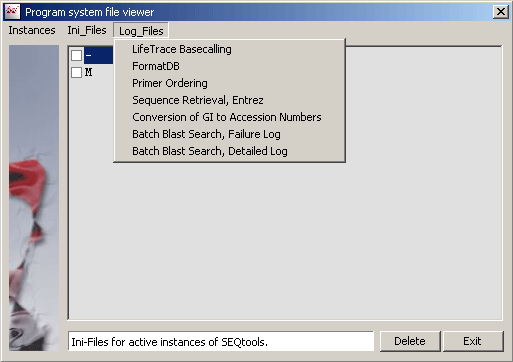
 4.14.10 log- and ini-file viewer
4.14.10 log- and ini-file viewer
Several log and data files are created by SEQtools or by the associated external programs. Often it is difficult to locate such files on your hard dish. This form includes links to all program generated text files.
 4.14.11 application color coding
4.14.11 application color coding
It is possible to run parallel instances of SEQtools. This can make it quite confusing/difficult to identify to which instance a particular form belongs. With the Color Coding option you can choose an identification color for each instance of SEQtools. This color is visible on most forms derived from the color coded insance.

© 2002-2010S.W. Rasmussen (revised: )