6. PROJECT SEQUENCE LIST
- 6.1 about the project sequence list
- 6.1.1 excluding sequences from list
- 6.1.2 sorting the sequence list
- 6.1.3 selecting data source for sequences
- 6.1.4 searching the sequence list
- 6.1.5 customising the displayed annotation
- 6.2 exporting the sequence list
- 6.2.1 export to new SEQtools instance
- 6.2.2 export to multi-sequence file
- 6.2.3 export to excel spreadsheet
 6.1 about the project sequence list
6.1 about the project sequence list
The project sequence list is one of the key features of seqtools designed to assist you in maintaining an overview of the often large number of sequences included in a project. The list displays a single information line for each sequence of the project - or for a selected range of project sequences.
The list form includes a number of options for customising the list. You can exclude sequences from the list on the basis of the quality of blast search results so only sequences with a significant match (expect value) to a particular database at genbank are included in the list.
The description lines returned from a genbank search can furthermore be formatted to remove irrelevant information - or information preventing a logical sorting of the description lines. The unformatted description is only a click away so you can easily check the original search result of the blast search.
The various options included in the sequence listing form are described in the following sections of this
page. The picture below shows how an fairly messy sequence list can be converted to a meaningful list of the positively classified project sequences.
Note that you always have the original blast search results at hand simply by highlighting a line and then holding down the right mouse button. This retrieves the blast section (shown below the list image) from the header for as long as you hold down the right mouse button.
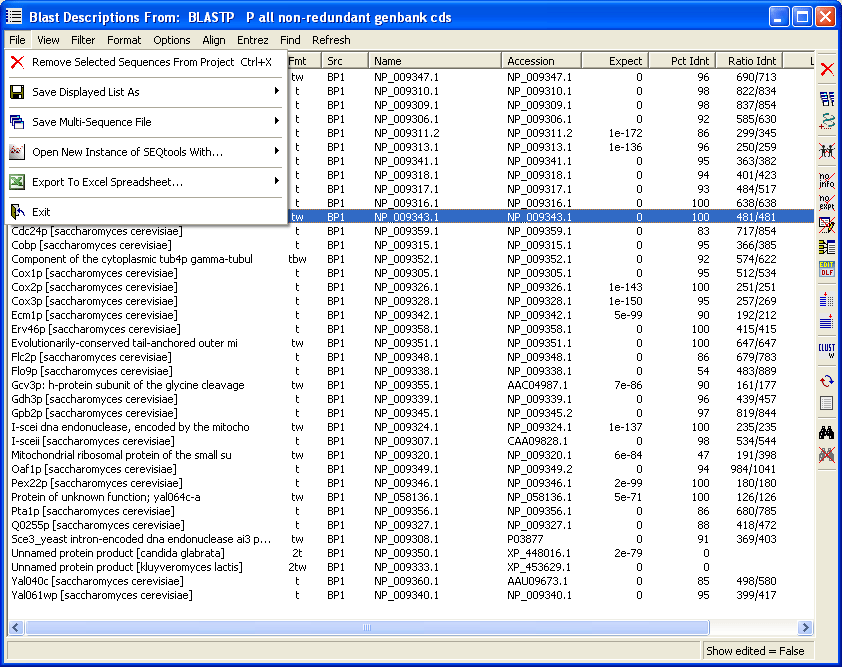
Once you have built the optimised sequence list the File menu includes several options for handling the included sequneces in the optimised list. The options are fairly obvious, except perhaps the possibility of exporting the information to an Excel spreadsheet. Using this option requires that Excel is availlable either already installed on your PC or downloaded and installed as part of seqtools. Use the form shown below to select which columns you wish to include in the spreadsheet file.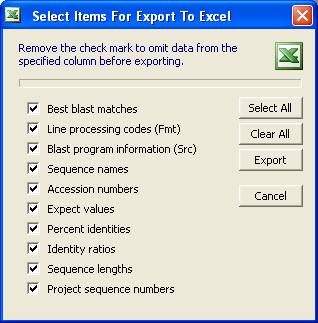
 6.1.1 excluding sequences from list
6.1.1 excluding sequences from list
There are a number of ways you can exclude project sequences from the list as shown on the unfolded Filter menu. You can hide sequences is they exists in more than one copy. The total number of copies is included duplicates in the Fmt column either as percent of all sequences in the project or as the actual number of copies of the sequence. Sequences with blast information as well as sequences where the blast expect value is outside the selected cutoff value.
If;
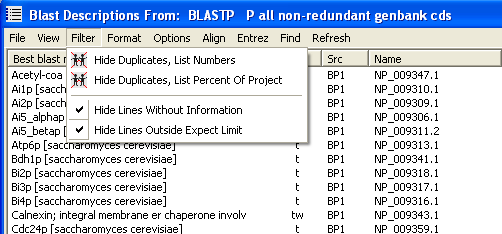
 6.1.2 sorting the sequence list
6.1.2 sorting the sequence list
You can sort the sequence list by clicking the column caption for each column. Each click toggles between descending and ascending sorting. The only exception is the blast expect value, The program is written in the programming language Visual Basic which treats the content of each column as strings. This is fine for columns including textual information but not correct for the numerical expect values. I have not yet found a method to get around this minor problem so you have to live with this inconvenience.
 6.1.3 selecting data source for sequences
6.1.3 selecting data source for sequences
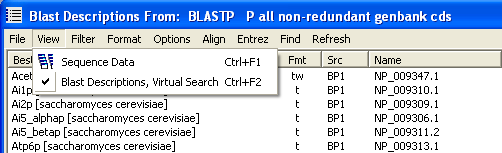
Depending on the search data available for the sequences the View menu will contain a varying number of options. In the project used to illustrate this section of the help file only blastP data are at hand so the menu contains the plain sequence date option and the blast descriptions from the search.
You use the Header Display Options form to compose the socalled virtual header wich is simply a temporary header including the blast data you have deciided to include. The form is shown below together with a form from a different project with more complete blast information available.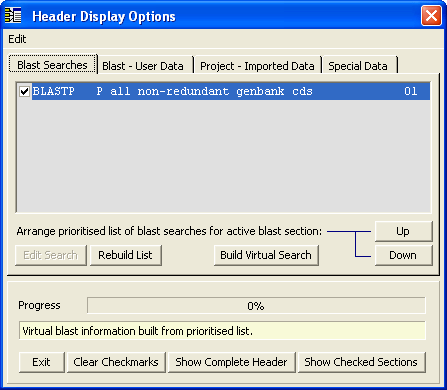
In this nucleotide project blast searches on the nr database have been conducted in addition to a blastX search om the complete protein database, tblastX on the yeast nucleotide database, and blastx searches on both D.melanogaster and E. coli translations. This project is the sample nucleotide project which can de
downloaded from the seqtools website.
Note that you need to select both the search data set and which part of the complete header information you wish to include in the virtual header. Look under the different tabs of the form to see the options. When you are done simply click Build Virtual Search to combine the selected information into the virtual header. Having built the virtual header you can choose to display the sequence list based on the information originating from each of the performed blast searches.
 6.1.4 searching the sequence list
6.1.4 searching the sequence list
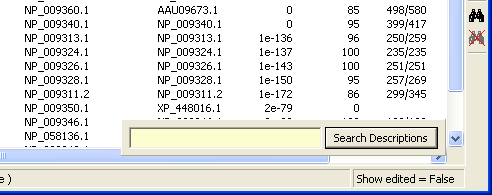
Pressing F3 displays the Search Descriptions search field which allows you to find entries in the sequence list. Note that the search starts from the beginning of the description line - and finds all occurrences of the query string. It is thus not possible to find a single word in the middle of a description line.
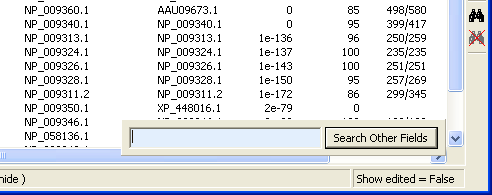
Clicking the F3 key a second time changes the reasch target to all other fields of the sequence list. This function will probably be modified in the future to improve the search process.
 6.1.5 customising the displayed annotation
6.1.5 customising the displayed annotation
There are a number of ways you can format the description lines to facilitate sorting and viewing the sequence data. How to build the virtual sequence header has already been described above. It is important to realise, that this is a necessary operation before beginning optimising the sequence list as this defines the data being available for the list.
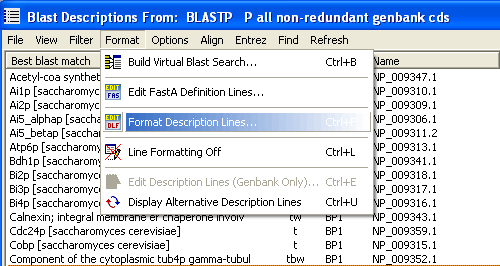
The key form for formatting description lines is shown and explained below. The first tab includes general options for description line formatting such as switching on and off the two truncate functions and the line replacement function, adjusting upper/lower case display, neglecting the best blast match using the second best match instead, a convenient options when search data originates from an local blast search where the best match is to the sequence itself. Also the blast expect value cutoff used in the sequence listing is set here.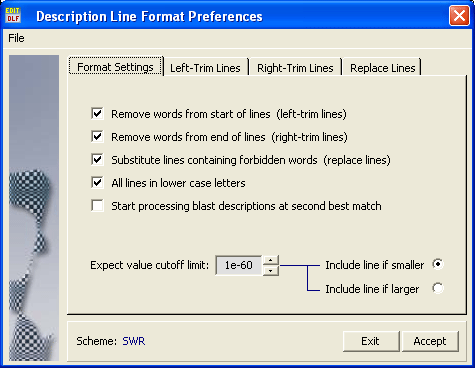
As shown in the examples below the description line returned from the database sometimes prevent proper sorting according to gene names. By removing the accession number and data base name from the beginning of the description lines solves this problem allowing you to perform a meaningful alphabetic sorting of the descriptions. It is possible to perform line truncation
either from the beginning or the end of the description line.
In cases where the description line refers to a hypothetical protein, an unknown protein, a putative protein etc... you can create a replacement list which causes extraction of the description line from the blast section to skip lines including one of the listed words. The function scans all descriptions until it finds a description line within the cutoff limit that does not contain any of the forbidden words. If a meaningful description line is not found, the first - and best - match is returned.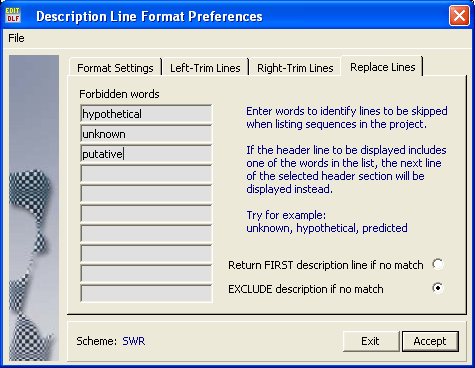
You can save your description line prosessing preferences in a *lfs file so they can be jused again.
Below are two examples showing the top of a sequence list before and after
left-truncating the description lines. The first showing the list before left-truncation has been performed.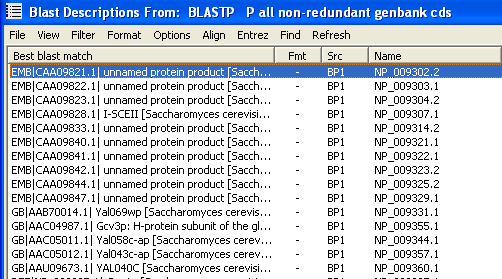
The description lines sorted alphabetically after left-truncation.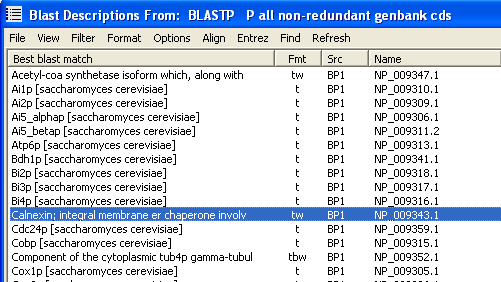
 6.2 exporting the sequence list
6.2 exporting the sequence list
As indicated in the beginning of this section of the seqtools manual there are a number of options for exporting the information included in the sequence list. The options are briefly described below.
 6.2.1 export to new seqtools instance
6.2.1 export to new seqtools instance
If you intend to continue working with the sequences included in the optimised list in seqtools, simply use this option to launch a new instance of seqtools which will contain all the sequences from your optimised list.
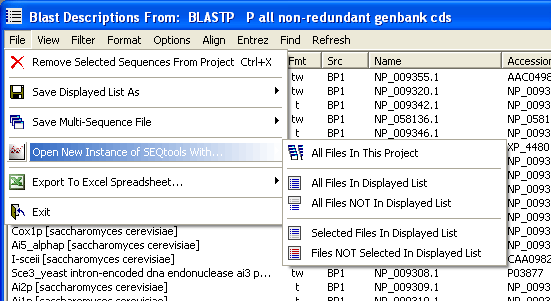
 6.2.2 exporting to multi-sequence file
6.2.2 exporting to multi-sequence file
The sequences included in the sequence list can be saved as a multi-sequence file which can be opened later in seqtools. The options available are evident from the image below.

 6.2.3 exporting to excel spreadsheet
6.2.3 exporting to excel spreadsheet
It has already been mentioned that the information included in the sequence list can be exported to an Excel spreadsheet. Note that you need to have Excel installed on your PC or to download and install the seqtools download file including excel.exe.

The options for selecting data for export to an Excel spreadsheet are as shown below.
© 2002-2010S.W. Rasmussen (revised: )
